Journal.pgen.0030185.g008.png
Summary
| Description Journal.pgen.0030185.g008.png |
English:
Native American
autosomal
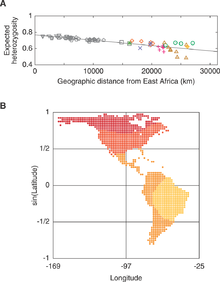
genetic tree. Each language stock is given a color, and if all populations subtended by an edge belong to the same language stock, the clade is given the color that corresponds to that stock. Branch lengths are scaled according to genetic distance, but for ease of visualization, a different scale is used on the left and right sides of the middle tick mark at the bottom of the figure. The tree was rooted along the branch connecting the Siberian populations and the Native American populations, and for convenience, the forced bootstrap score of 100% for this rooting is indicated twice.
In the neighbor-joining tree, a reasonably well-supported cluster (86%) includes all non-Andean South American populations, together with the Andean-speaking Inga population from southern Colombia. Within this South American cluster, strong support exists for separate clustering of Chibchan–Paezan (97%) and Equatorial–Tucanoan (96%) speakers (except for the inclusion of the Equatorial–Tucanoan Wayuu population with its Chibchan–Paezan geographic neighbors, and the inclusion of Kaingang, the single Ge–Pano–Carib population, with its Equatorial–Tucanoan geographic neighbors). Within the Chibchan–Paezan and Equatorial–Tucanoan subclusters several subgroups have strong support, including Embera and Waunana (96%), Arhuaco and Kogi (100%), Cabecar and Guaymi (100%), and the two Ticuna groups (100%). When the tree-based clustering is repeated with alternate genetic distance measures, despite the high Mantel correlation coefficients between distance matrices (0.98, 0.98, and 0.99 for comparisons of the Nei and Reynolds matrices, the Nei and chord matrices, and the Reynolds and chord matrices, respectively), higher-level groupings tend to differ slightly or to have reduced bootstrap support.
|
| Date | |
| Source |
Source: Genetic Variation and Population Structure in Native Americans- Published online 2007 November 23. doi: 10.1371/journal.pgen.0030185
PUBLIC LIBRARY OF SCIENCE (Original Map site): [1] |
| Author | Tamm et al. |
|
Permission
( Reusing this file ) |
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. The Public Library of Science (PLoS) applies the Creative Commons Attribution License (CCAL) to all works. ( http://www.plosone.org/static/license.action ) |
| Other versions |
|
Licensing
This file is licensed under the
Creative Commons
Attribution 2.5 Generic
license.
-
You are free:
- to share – to copy, distribute and transmit the work
- to remix – to adapt the work
-
Under the following conditions:
- attribution – You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
|
|
This file, which was originally posted to
https://www.ncbi.nlm.nih.gov
, was reviewed on 16 August 2011 by
reviewer
Hoangquan hientrang
, who confirmed that it was available there under the stated license on that date.
|
Original upload log
The original description page was
here
. All following user names refer to en.wikipedia.
- 2010-02-02 21:08 Moxy 546×600× (82861 bytes) Each language stock is given a color, and if all populations subtended by an edge belong to the same language stock, the clade is given the color that corresponds to that stock. Branch lengths are scaled according to genetic distance, but for ease of visu
Captions
Add a one-line explanation of what this file represents

